We recently published two preprints that provide the strongest evidence yet that the COVID-19 pandemic began at a market selling live animals in Wuhan.
An almost mirror-image of SARS just two decades earlier.
zenodo.org/record/6291628
zenodo.org/record/6299600
Long 🧵 👇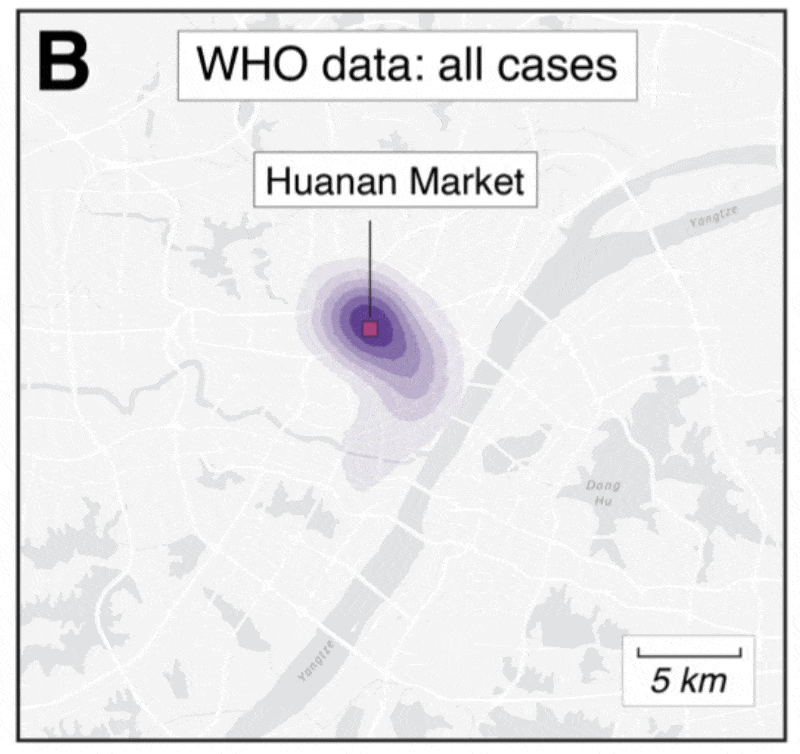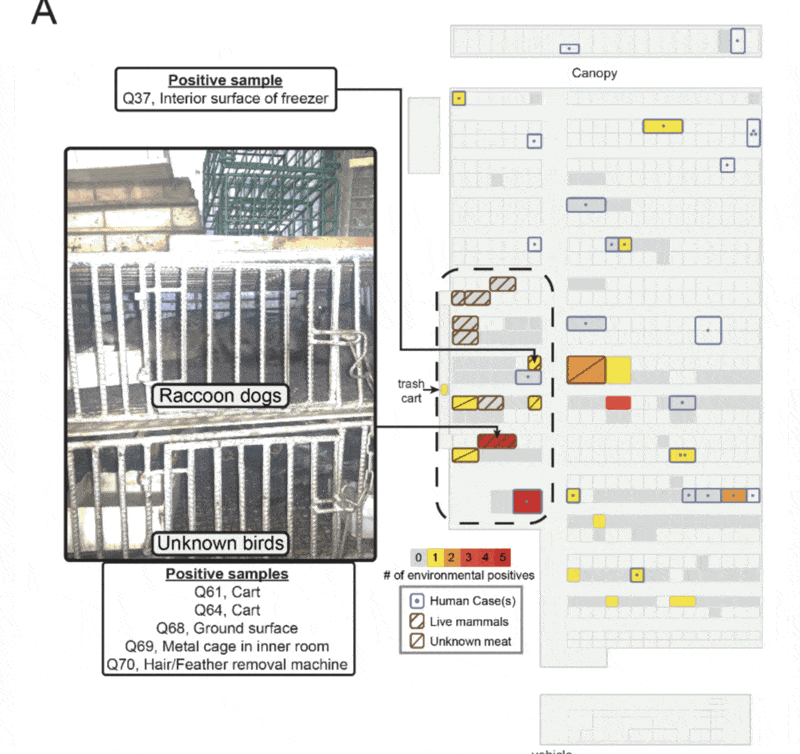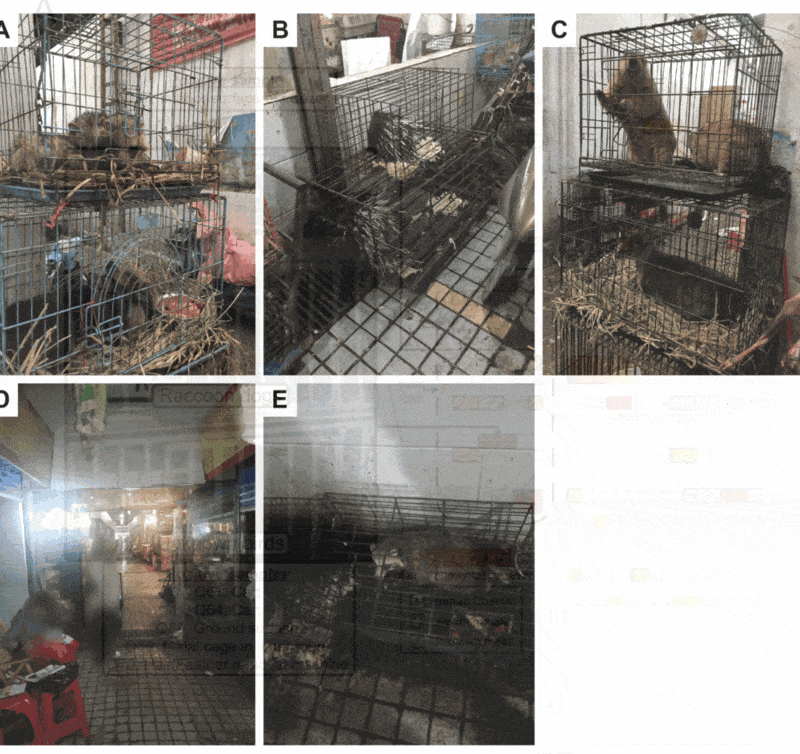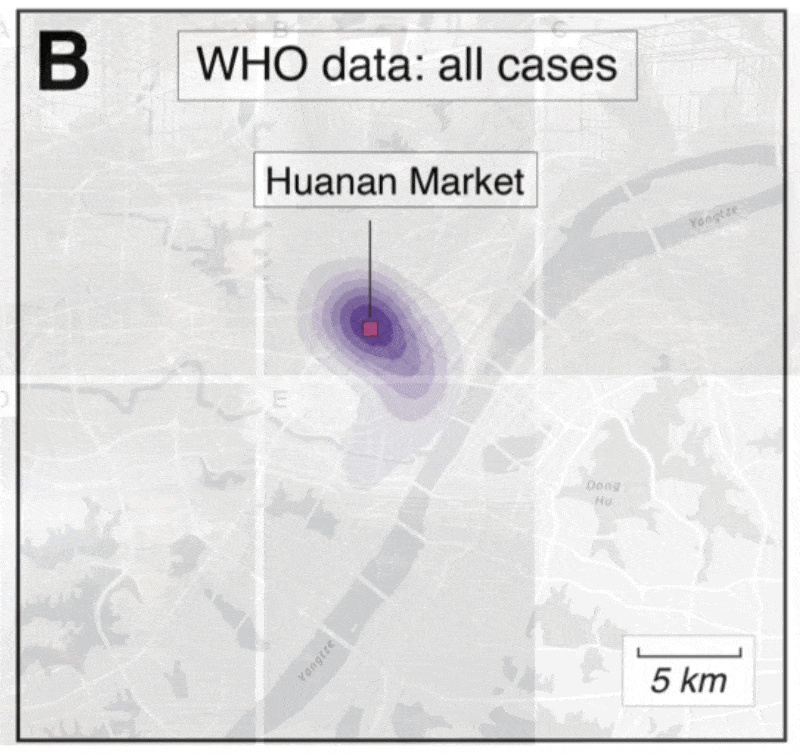
Why do we believe the Huanan market was the origin?
1️⃣ Precedent
2️⃣ Clustering of hospitalizations, cases, deaths
3️⃣ Not a superspreading event
4️⃣ Two introductions
5️⃣ Susceptible animals sold
6️⃣ Clustering where animals were sold
7️⃣ Animal-associated objects ➕
Details 👇
Before I dive in, I want to answer the most important question - does this mean we have solved the origin of the pandemic?
No, we have not. It is clear that it began at a market, but most everything upstream remains a black box and there's still a ton to do.
More at the end 🧵.
📖 In the first preprint, led by @jepekar we show that:
"SARS-CoV-2 emergence very likely resulted from at least two zoonotic events"
zenodo.org/record/6291628
📖 In the second preprint, led by @MichaelWorobey we show that:
"The Huanan market was the epicenter of SARS-CoV-2 emergence"
zenodo.org/record/6299600
1️⃣ Precedent
The emergence of SARS-CoV-2 looks a lot like that of SARS-CoV-1, especially:
✅ Virus similarity (both are sarbecoviruses)
✅ Timing (November 2002 vs November 2019)
✅ Host range (broad)
✅ Association with wet markets
For example, compare...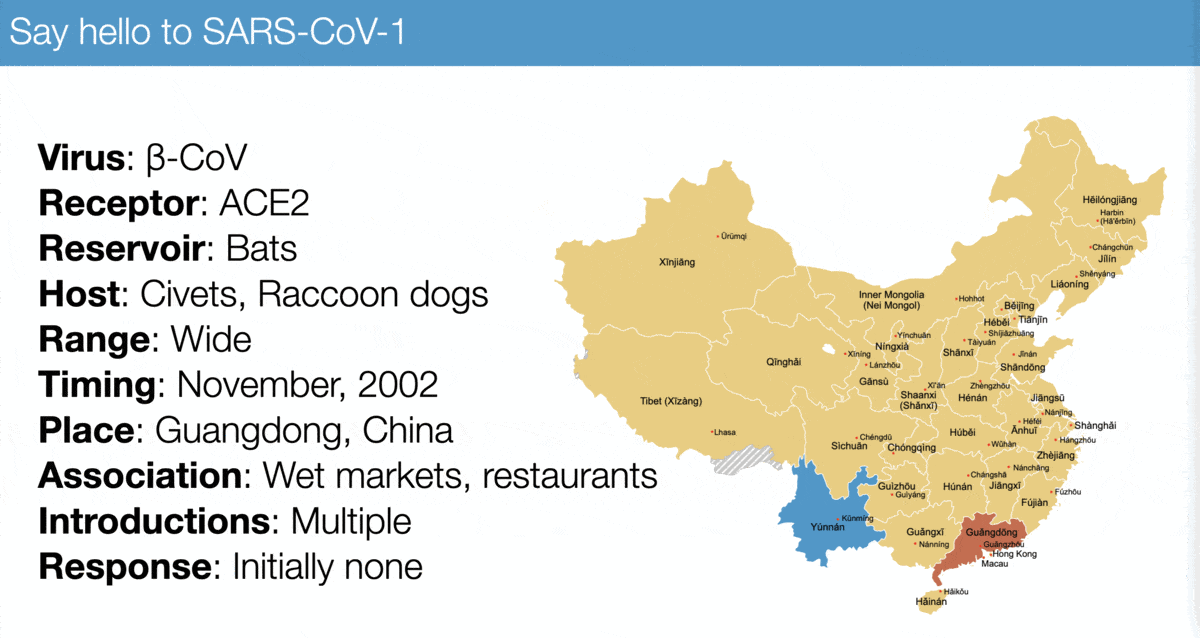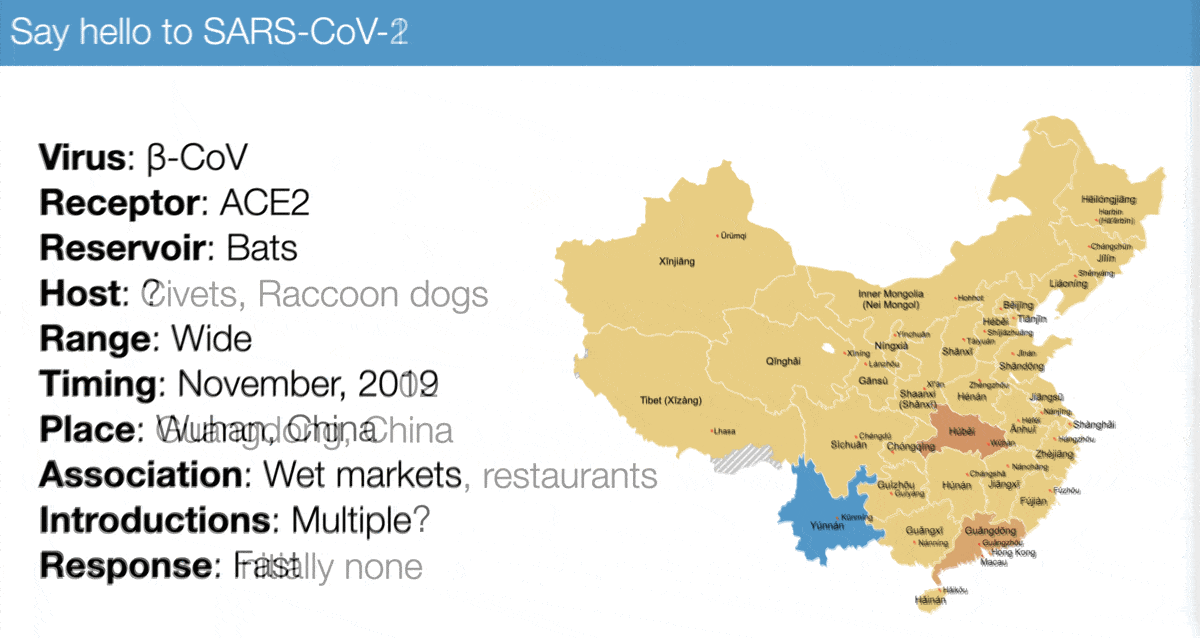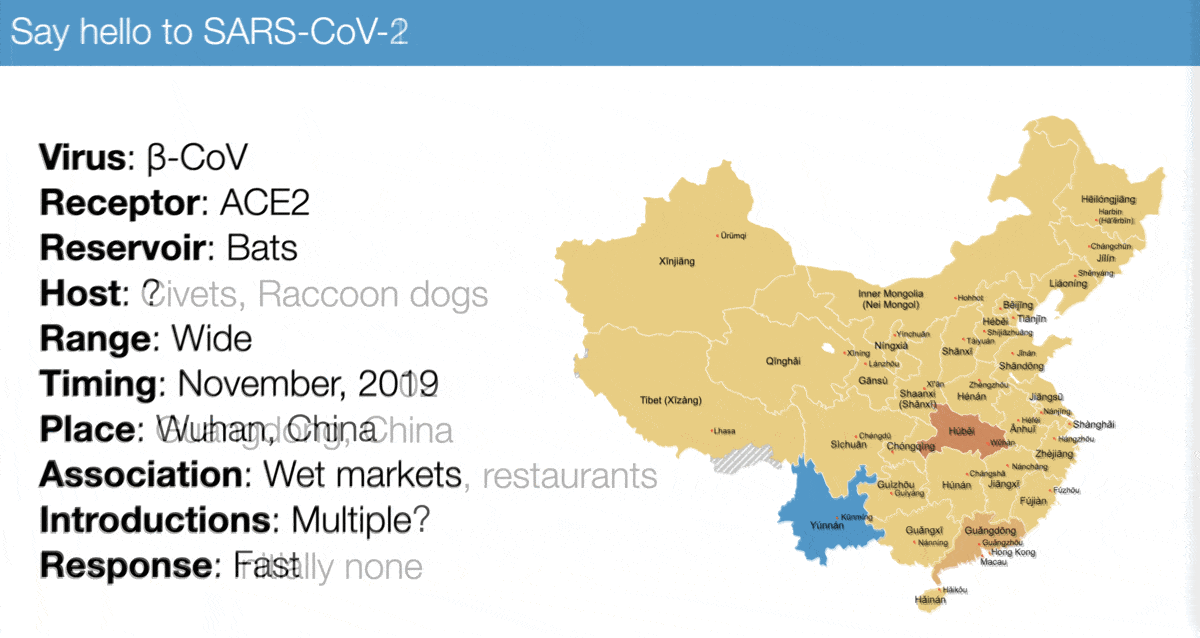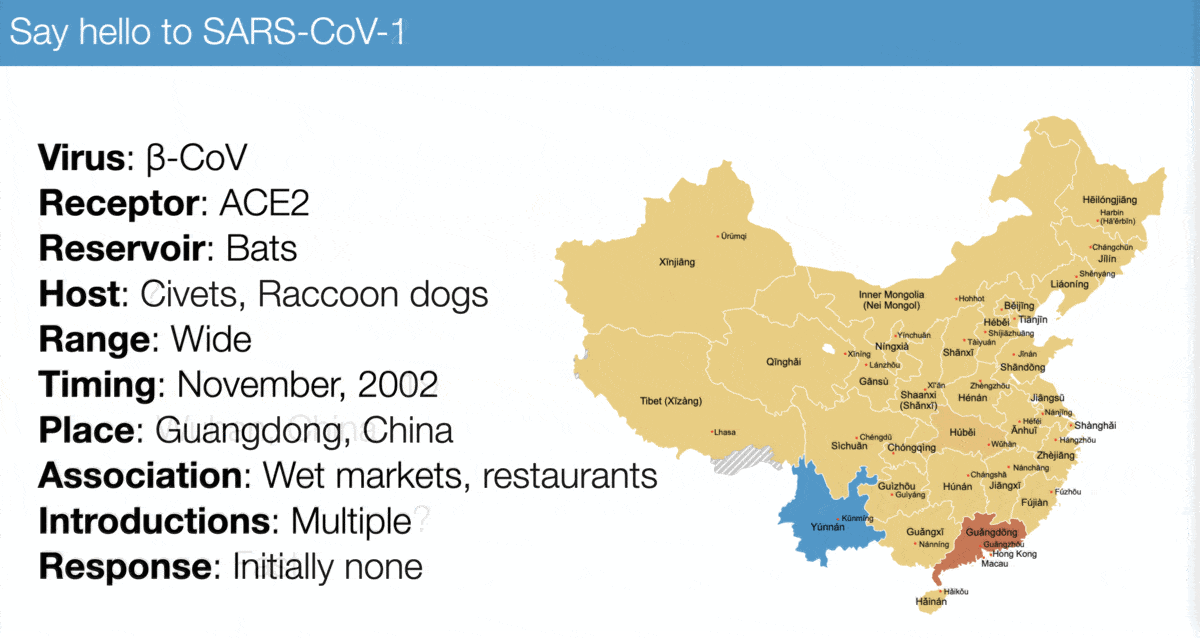
The emergence of a novel coronavirus isn't exactly news - it happens frequently. Here's a sampling of the ones we know about 👇 .
And, of course, since the beginning of the pandemic, many closely related viruses have been found, including BANAL-52:
nature.com/articles/s41586-022-04532-4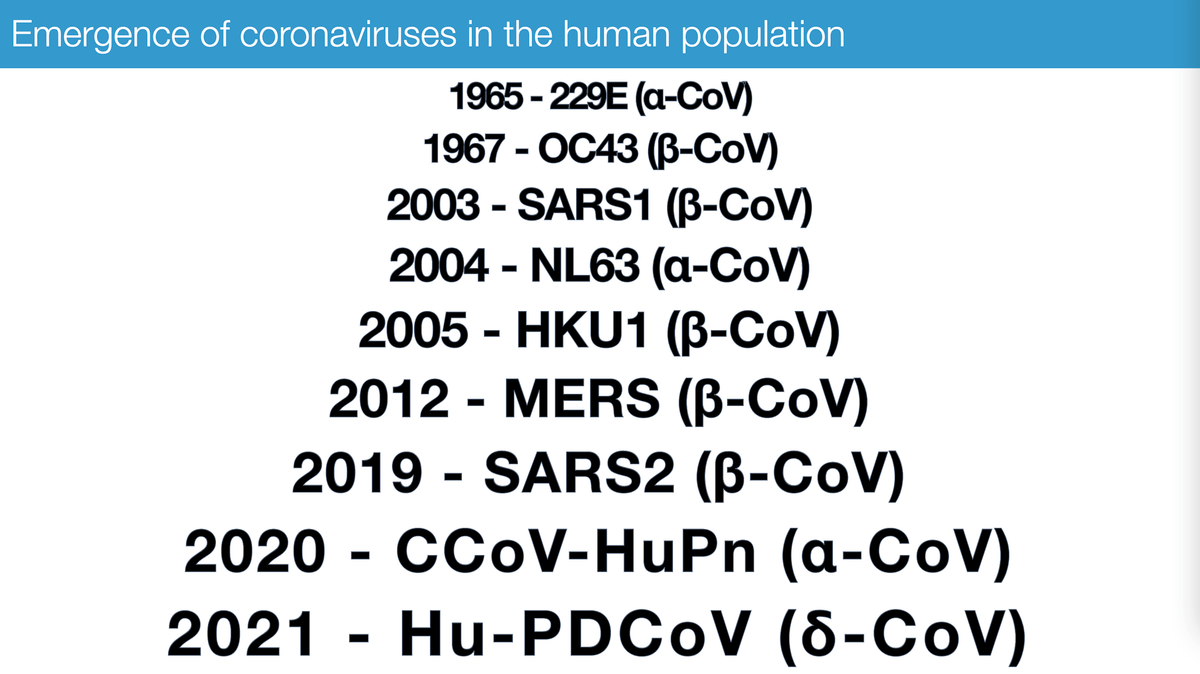
These viruses are widespread all across South-East Asia because their reservoirs - horseshoe bats - are widely distributed, including, yes, in Hubei province. Importantly, virus diversity is massively undersampled.
Excellent paper led by @SpyrosLytras:
science.org/doi/10.1126/science.abh0117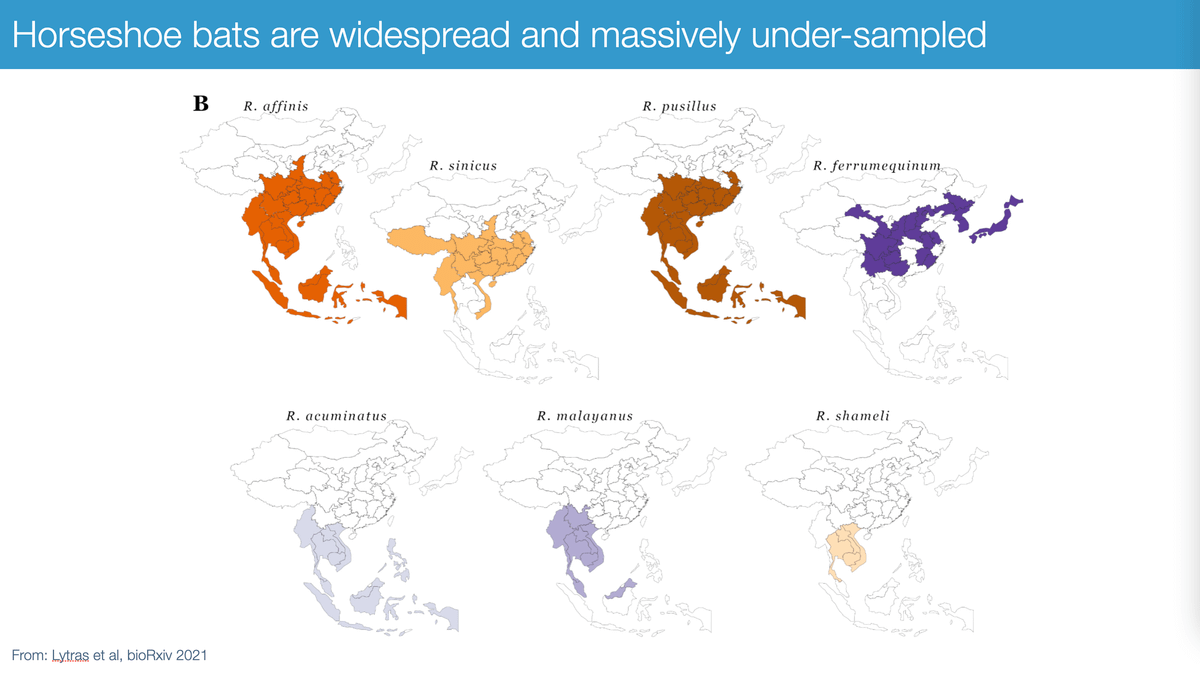
If we go back to SARS1, we know that infected animals were found on farms in Hubei in both 2003 and 2004 - outside Wuhan. In fact, more SARS1 infected animals were found on Hubei farms than anywhere else in China - and Hubei supplied animals to Guangdong.
twitter.com/K_G_Andersen/status/1488918972003094529?s=20&t=Lc1K_fAPVgT9VdVW8W-aIw
Interestingly, the SARS1 genomes from Hubei - unlikely those from Guangdong - are significantly closer to the SARS1 genomes from human patients.
Were Hubei farms the potential source for SARS1 and SARS2? Maybe, but need much more research.
References:
twitter.com/K_G_Andersen/status/1488718228238974983?s=20&t=j-VcqV9tc-t9wMEEpVHFeg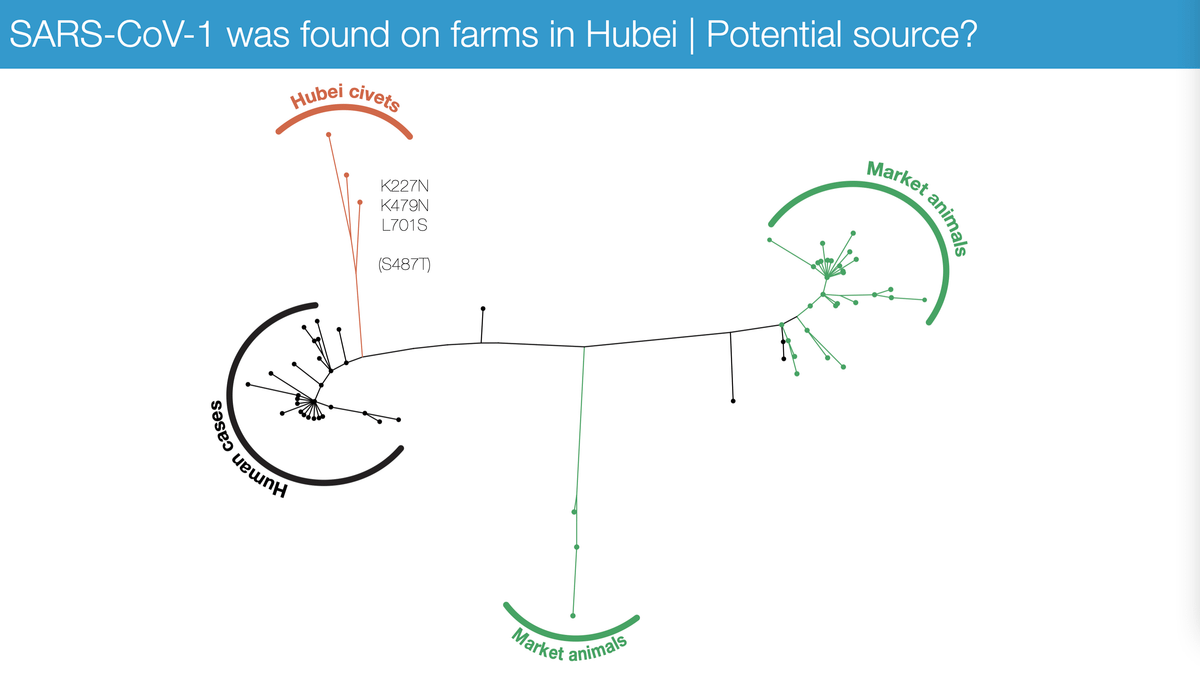
Because SARS-CoV-2 became a pandemic virus and SARS-CoV-1 did not, we would expect to find some 'key' features in this virus. We discussed this in our "Proximal Origin" paper:
nature.com/articles/s41591-020-0820-9
And this review:
cell.com/cell/fulltext/S0092-8674(21)00991-0
I previously did a lengthy thread on one of these features - the furin cleavage site. Some have suggested that the structure of this site is suggestive of engineering - that couldn't be further from the truth (quite the opposite).
twitter.com/K_G_Andersen/status/1391507230848032772
So why Wuhan? Another thread from @MichaelWorobey:
twitter.com/MichaelWorobey/status/1438202006678630404
2️⃣ Clustering of hospitalizations, cases, deaths around the Huanan market
@MichaelWorobey wrote an excellent perspective on the clustering of hospitalizations early in the pandemic that showed a *very* clear association to the Huanan market:
science.org/doi/10.1126/science.abm4454
I previously talked about the clustering of cases and excess deaths when I did a (rather lengthy...) rundown of the @WHO mission report:
twitter.com/K_G_Andersen/status/1376954932004196352
In the first part of Worobey et al., we go much deeper on the "clustering" analysis:
✅ Early cases are clearly associated with the market
✅ Association is non-random and not a result of age/demographics
✅ The Huanan market was the early epicenter
zenodo.org/record/6299600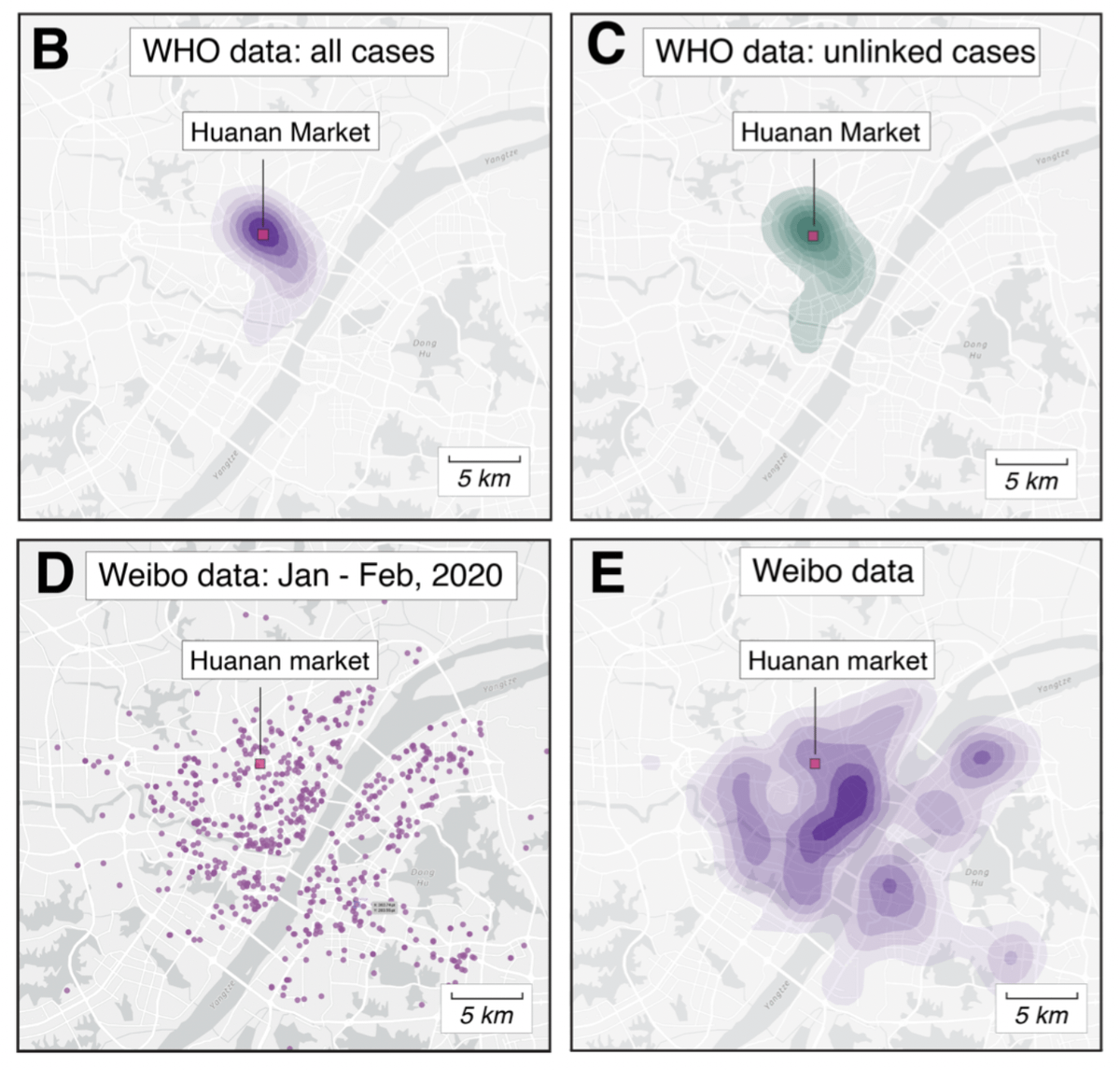
We extracted early case locations from the WHO mission report, later (probable) case locations from Weibo, and created null models based on population density or Jan/Feb cases.
No matter how we look at the data, the association is clear and non-random.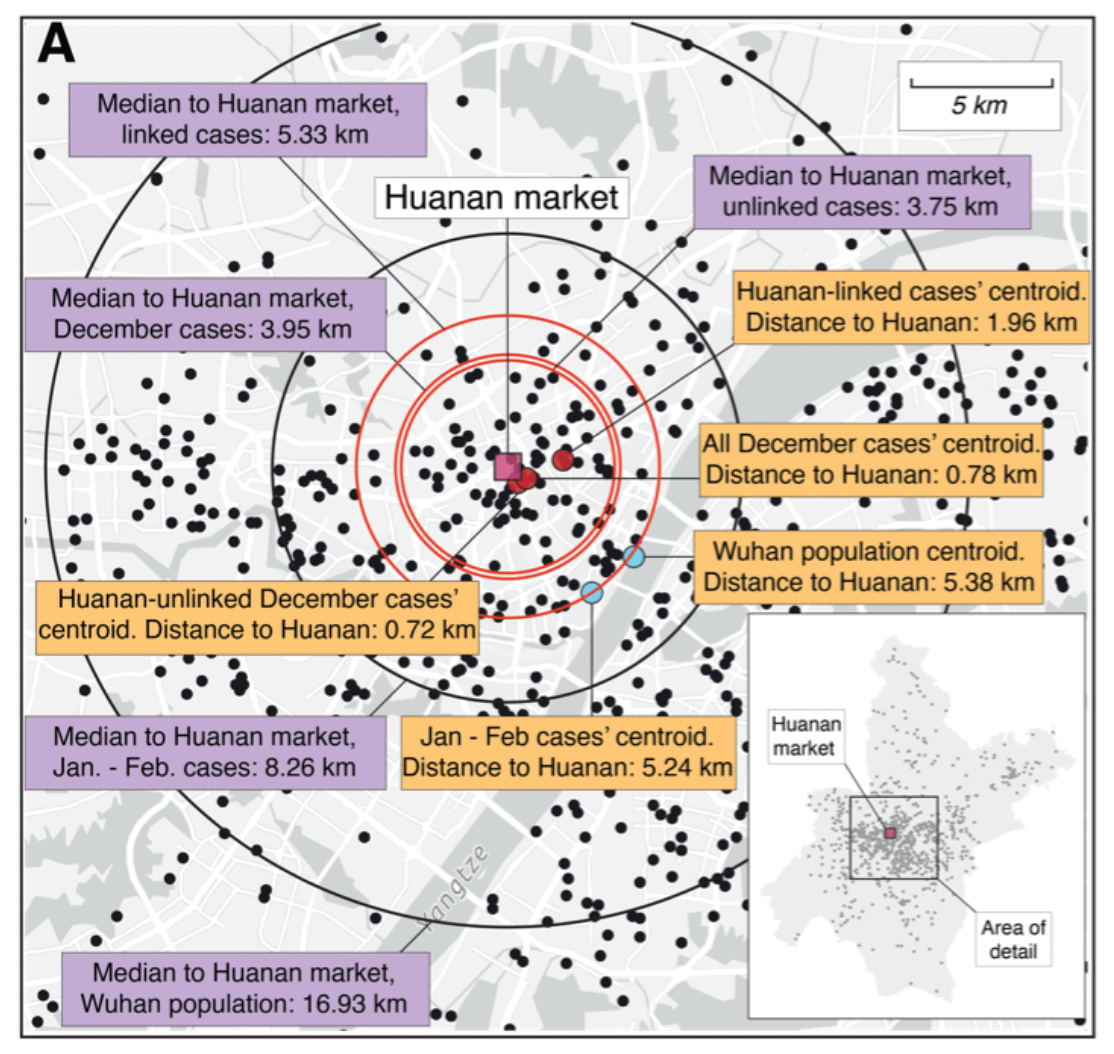
Further, the Huanan market is *the only* place in Wuhan where early cases had a clear association - there are no other epidemiological links to any other place in the city.
All of that changed as the outbreak spread - then we see clear association based on demographics.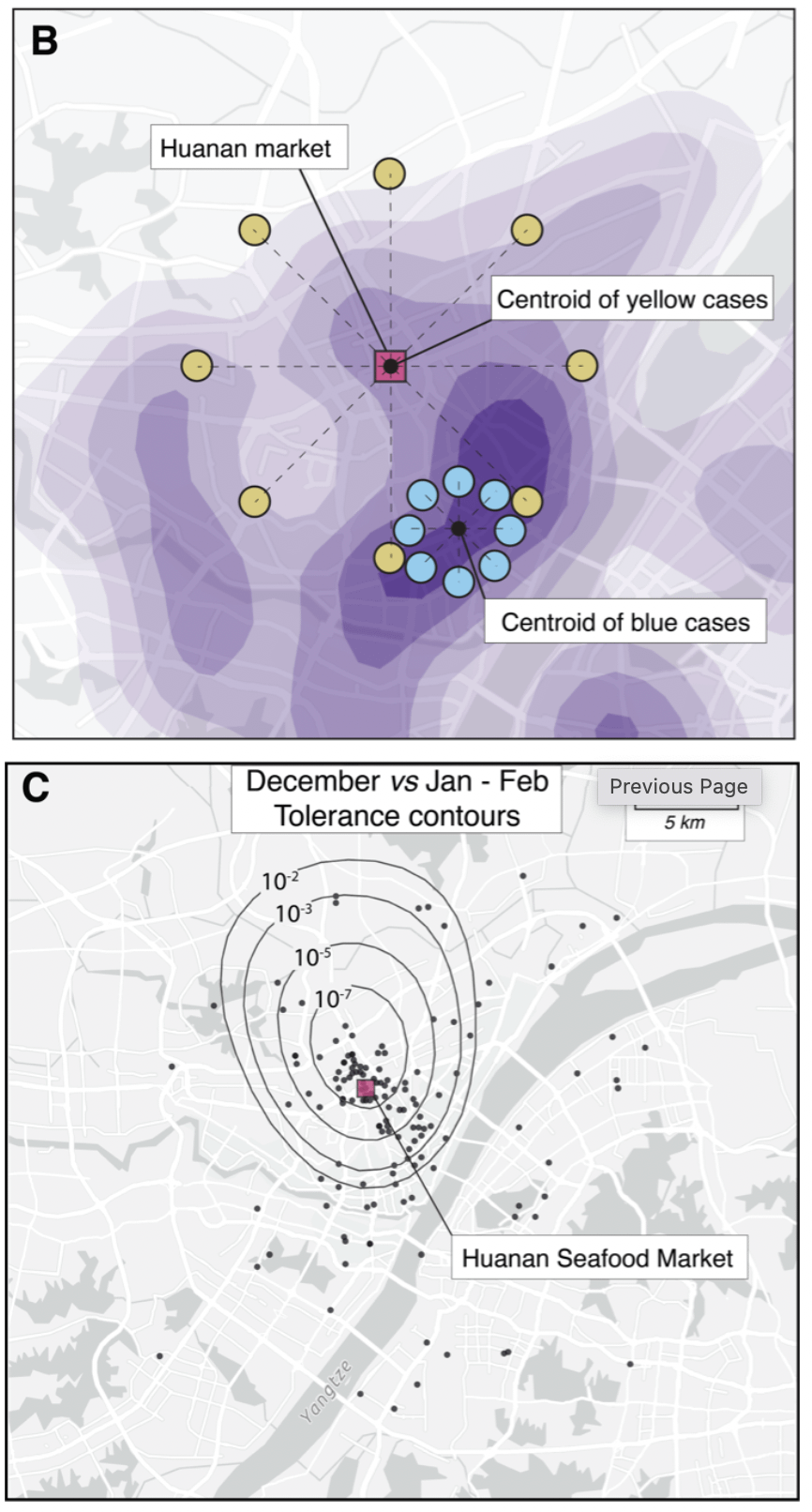
3️⃣ Not a superspreading event
Some have taken this to mean that the market was "merely a superspreading event". This is very unlikely:
✅ Clustering inside market
✅ Timing + multiple spillovers
Further, the doubling rate in the market was ~3-4 days (estimated from WHO data👇)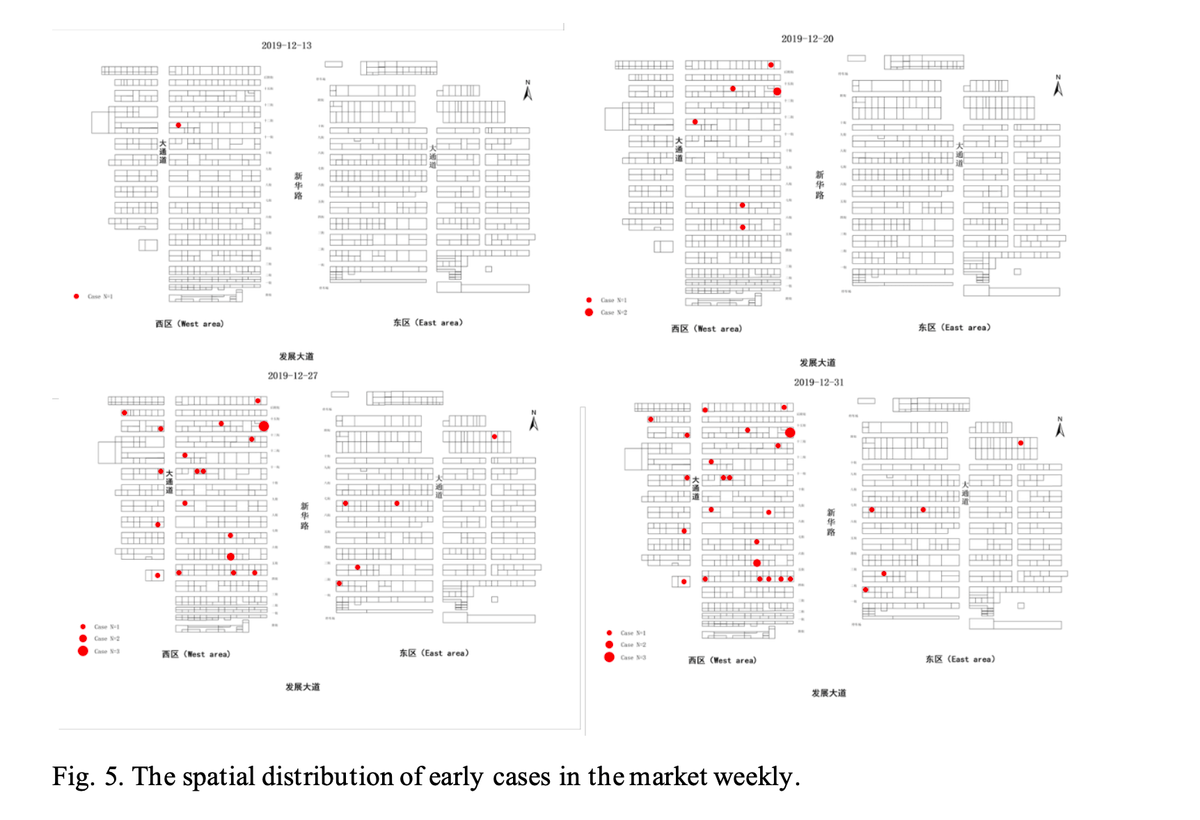
4️⃣ Two introductions
Let's hop over to Pekar et al., where we show that it's very likely that SARS-CoV-2 jumped multiple times (like SARS-CoV-1) - with one 'successful' lineage ("B") likely spilling over in late November and lineage "A" 1-2 weeks later.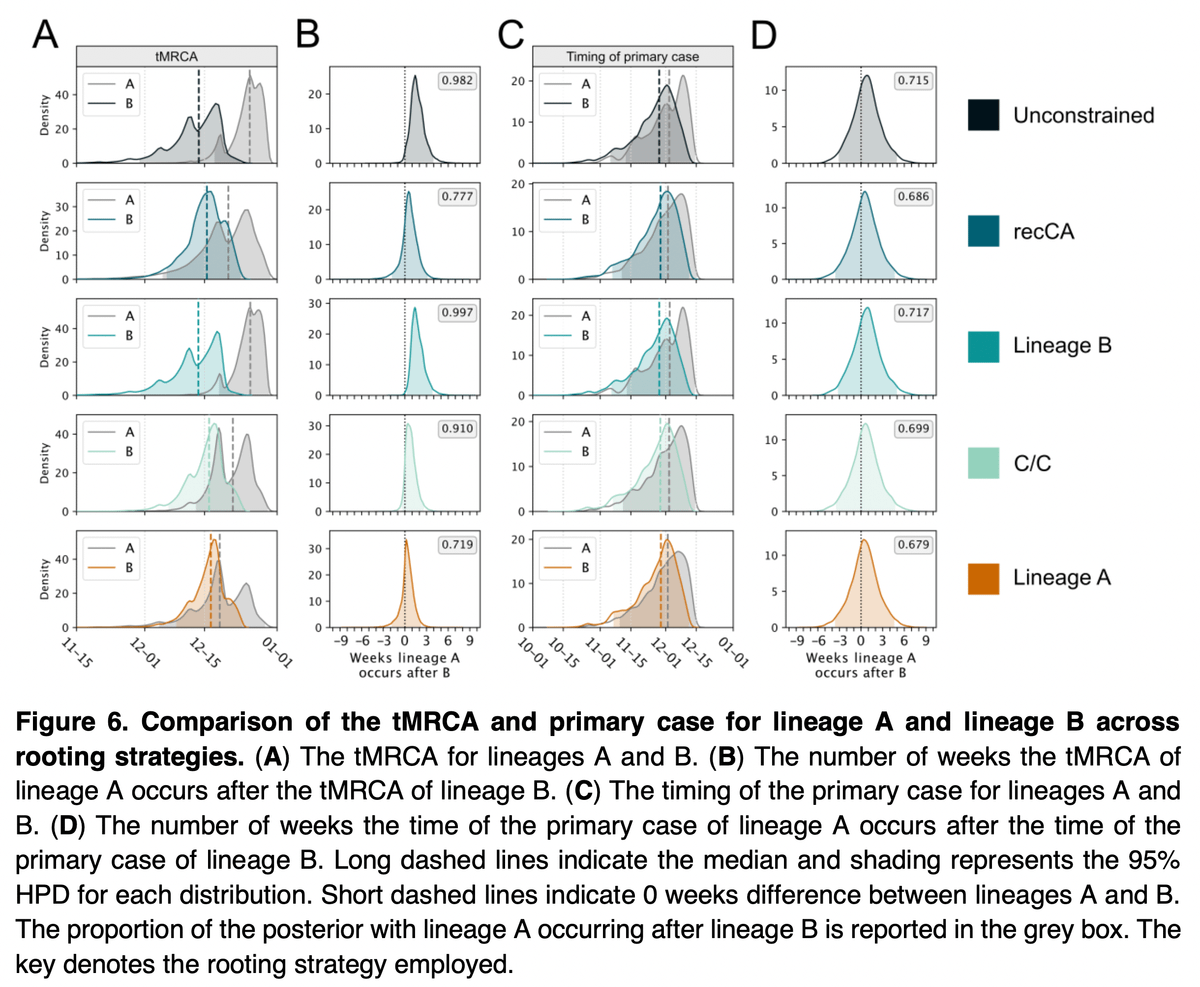
One quick comment on "multiple spillovers" as some people find this exceedingly unlikely ("one spillover is unlikely, so two spillovers are much more unlikely").
This is not correct. We're dealing with conditional probability - one happens, two (and more) are likely to happen.
As Joel Wertheim put it - "we failed to climb Mount Everest for hundreds of thousands of years. And then, in just one day, two people did".
This is the same. The "(un)likelihoods" are upstream of infected animals - with just the right virus - ending up in a market. Once done,💥
We have known for a while that two early lineages of SARS-CoV-2 existed - Lineage "A" and Lineage "B", using the PANGO naming convention.
These two differ at two sites - 8272 ("A"=T; "B"=C) and 28144 ("A"=C; "B"=T) so "A" is T/C and "B" is C/T.
zenodo.org/record/6291628
Genomic data show that "intermediates" (C/C; T/T) between A and B may have existed during the early outbreak in Wuhan - suggesting that e.g., A evolved to B or B evolved to A in humans.
However, we show that this likely wasn't the case - early intermediates are due to errors.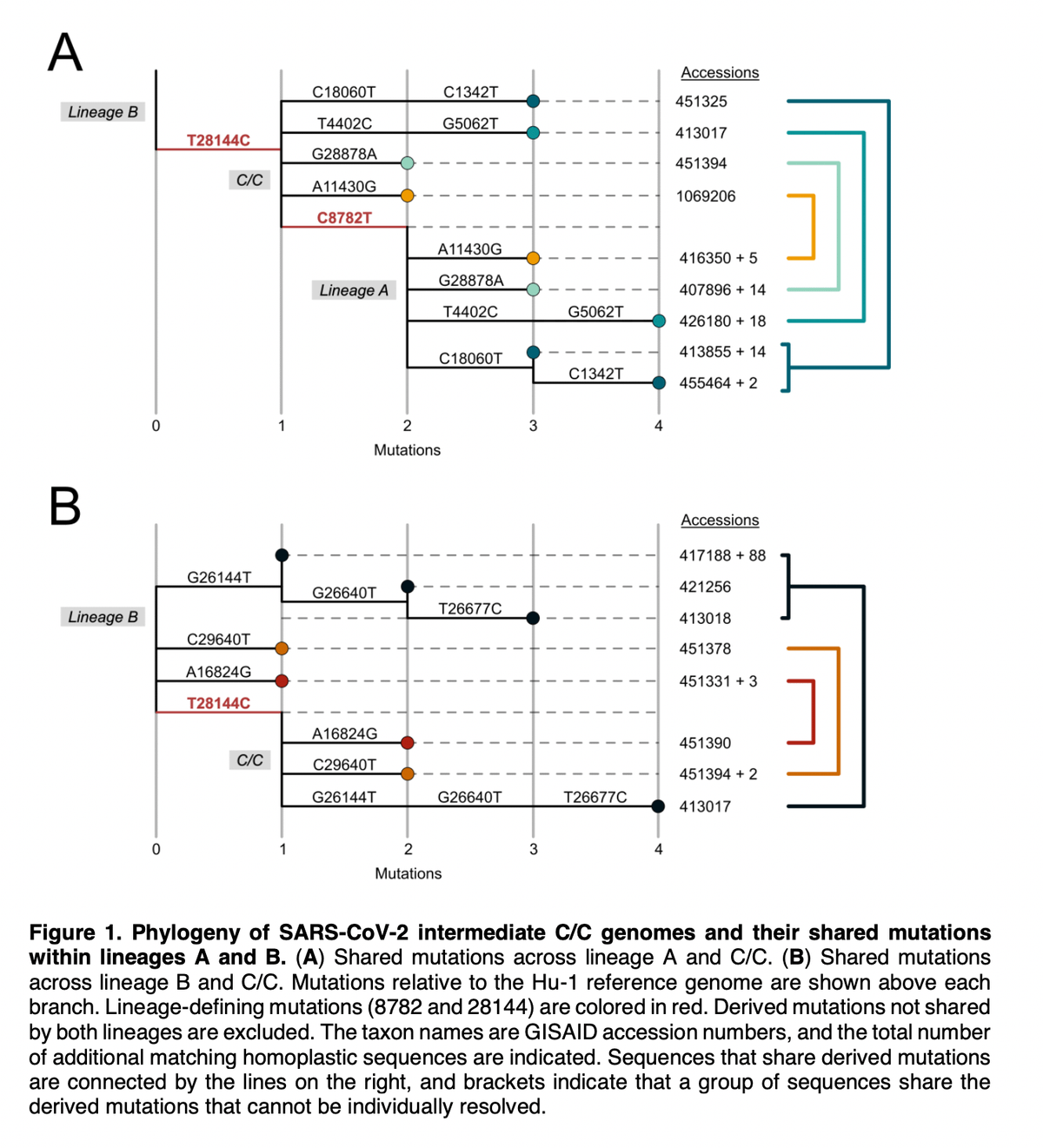
Lineage A is closer to closely related viruses like RaTG13 and BANAL-52, however, we can't simply say that this means that A is the ancestor of B as some have done previously.
E.g.,
academic.oup.com/mbe/article/38/12/5211/6353034
Trying to establish a rooted phylogeny of SARS-CoV-2 requires much more careful analysis than simply performing "outgroup" rooting (like ☝️).
In Pekar et al., we performed several Bayesian analyses to estimate likely roots and also created a putative "common ancestor".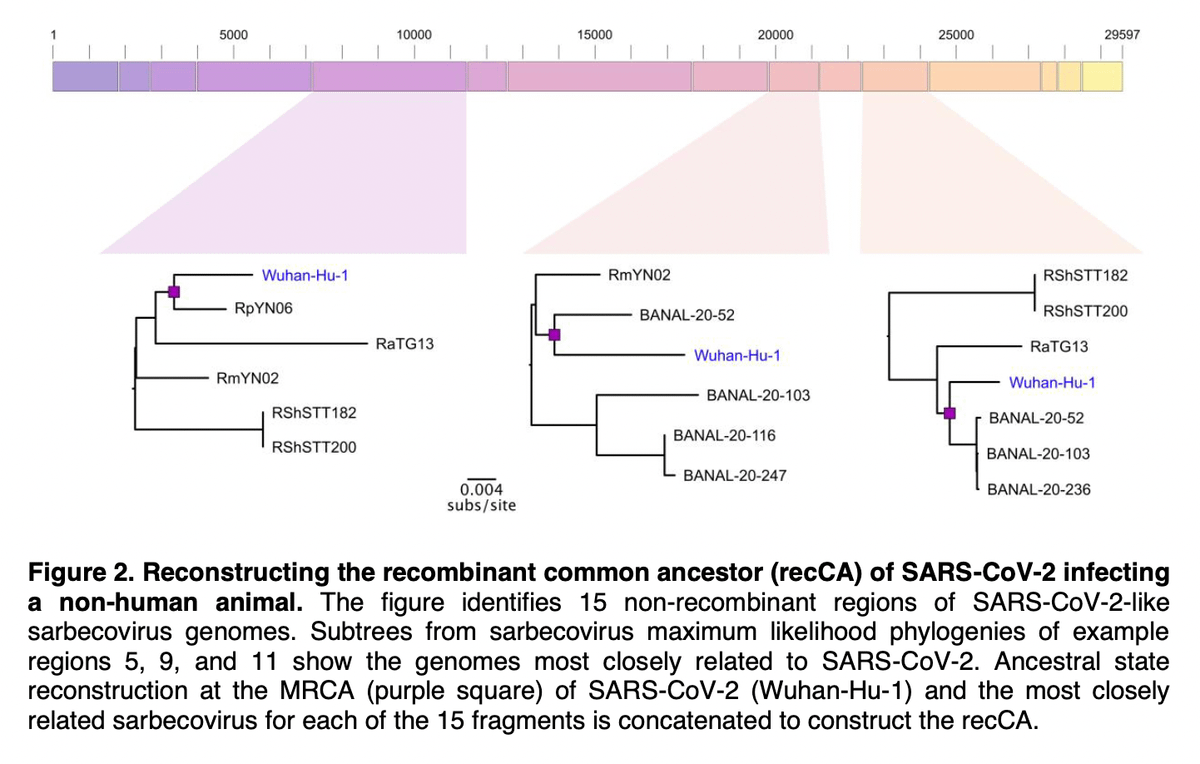
Once we obtain proper (posterior) estimates of the early SARS-CoV-2 phylogeny - and incorporate factors such as sampling times and evolutionary models - we can simulate plausible scenarios of early virus diversity and compare to the diversity that was actually observed.
These analyses make it clear that it is *much* more likely that A and B resulted from independent spillovers - one did not evolve into the other in the human population.
Further, our timing estimates show that B likely spilled over late-November and A a little after that.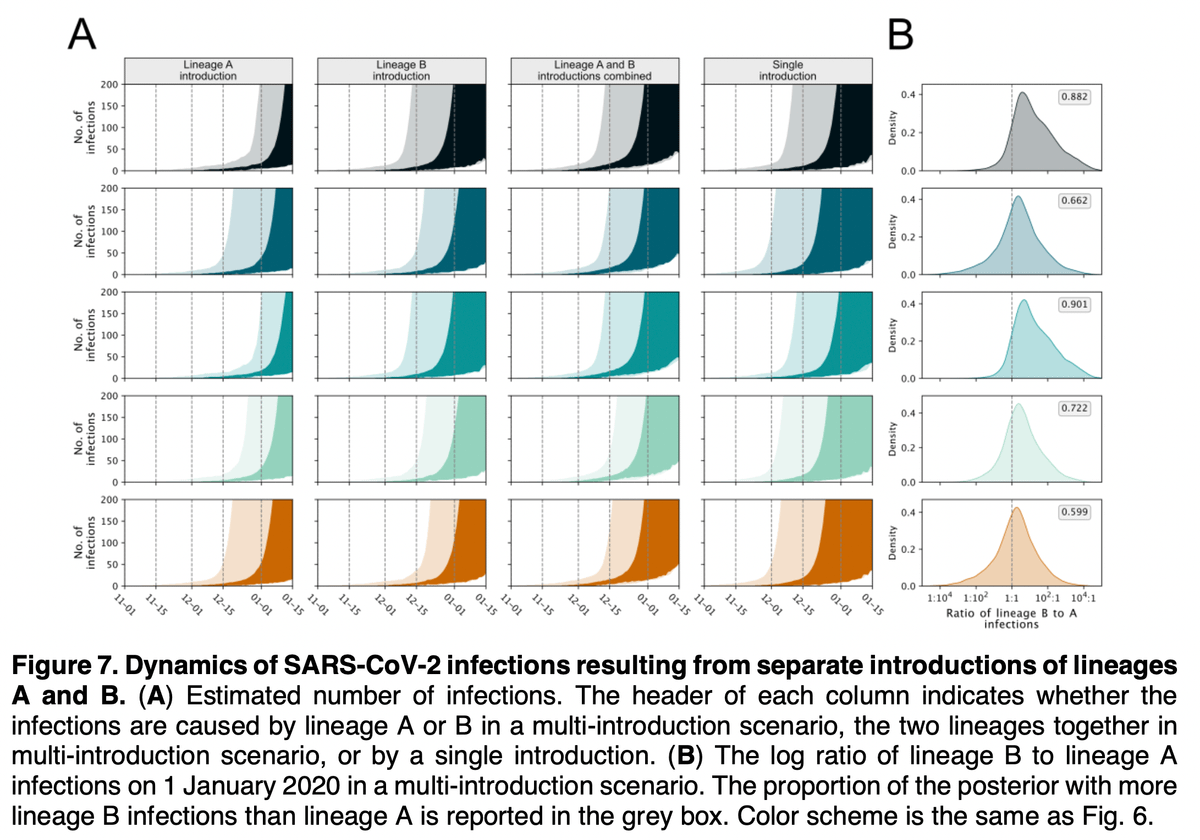
These findings need to be seen in the context of a clear geographical association of *both* Lineages A and B with the Huanan market.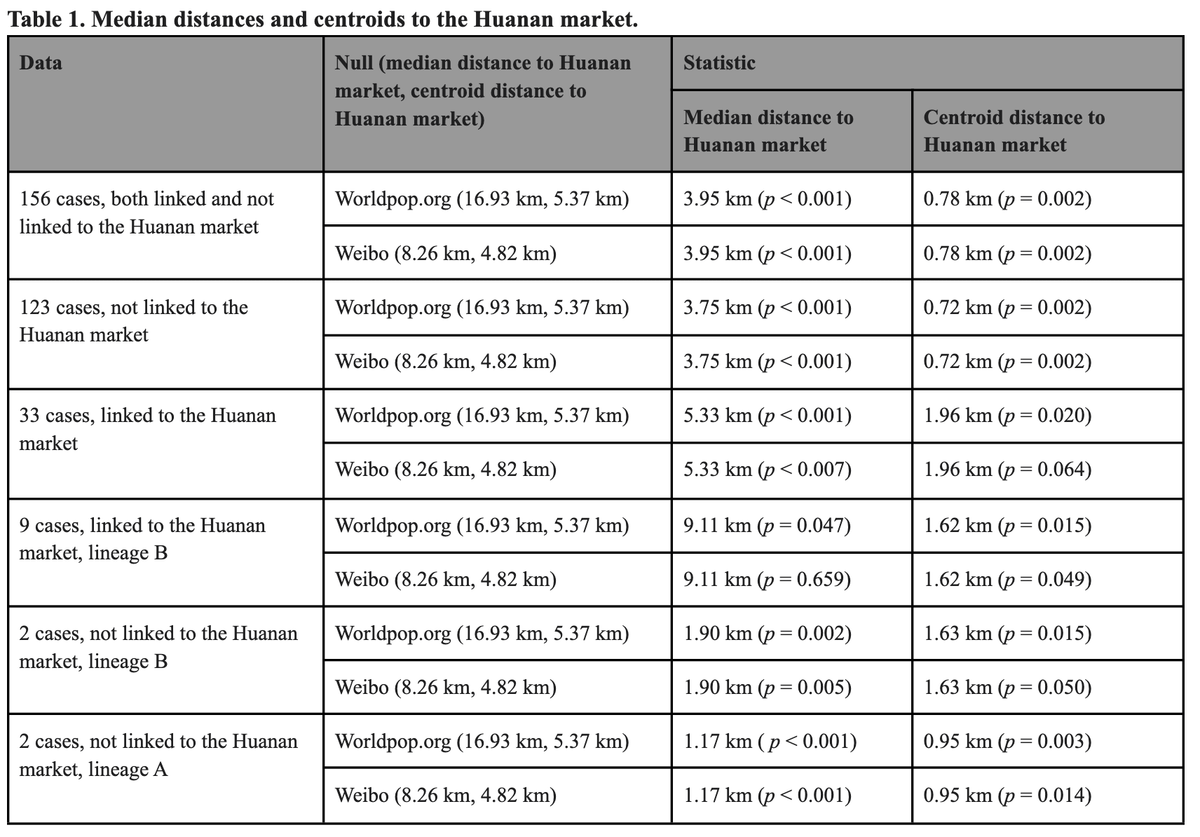
Further, as we were wrapping up our preprints, Gao et al. posted a preprint showing that Lineage A was indeed present at the Huanan market - in an environmental sample ("A20"), no less.
researchsquare.com/article/rs-1370392/v1
We also don't see any evidence of intermediate genomes at the Huanan market in the Gao et al. preprint. We can therefore conclude:
✅ Both Lineage A and B found at Huanan market
✅ Early intermediates likely not real
✅ Very likely two independent spillovers at the Huanan market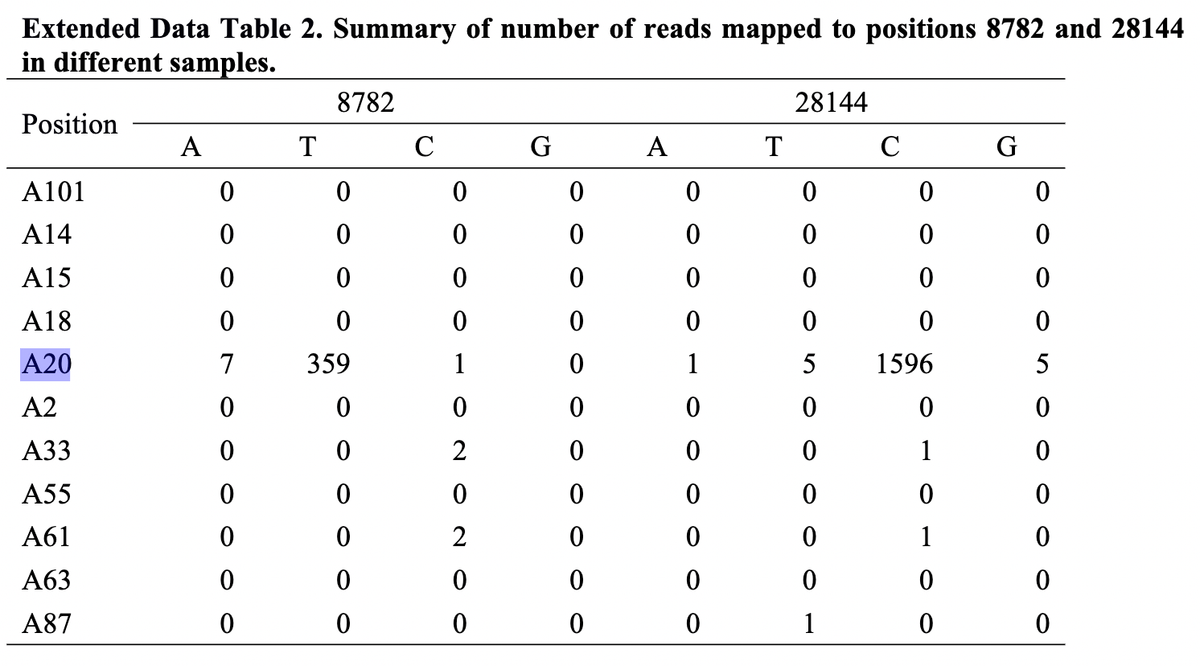
5️⃣ Susceptible animals sold at the Huanan market in Nov/Dec 2019
Having established a clear - undeniable - association of early cases to the Huanan market of early cases, we next investigated likely conduits.
We already knew live animals were sold 👇
nature.com/articles/s41598-021-91470-2
But what was on sale during the critical months of November and December? Well, same types of animals described in the Xiao et al. preprint, including raccoon dogs.
Based on surveys co-author Chris Newman was part of, we had data for November: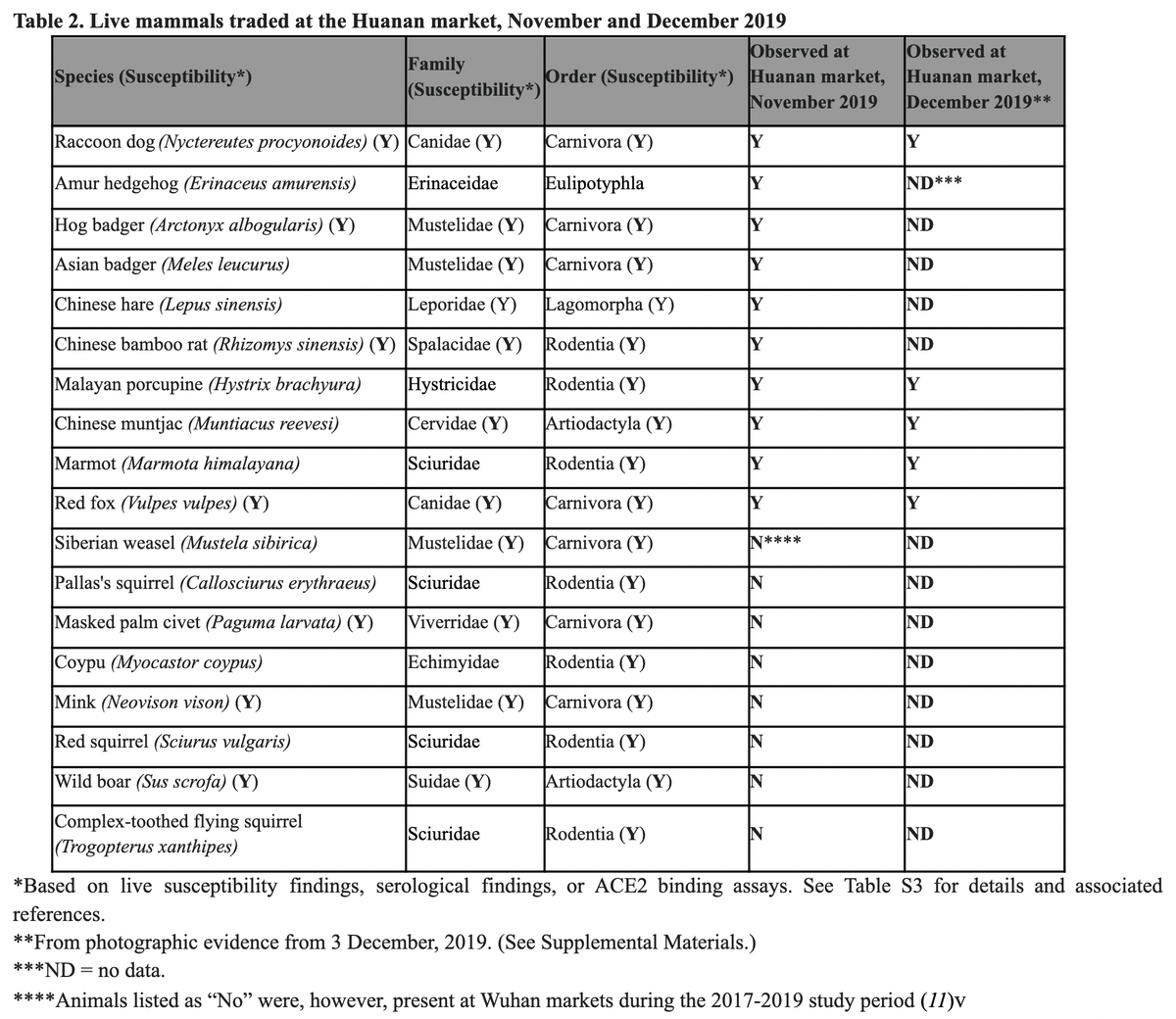
And based on publicly available images and recordings, we know e.g., raccoon dogs were also on sale in December.
youtube.com/watch?v=Je0_U2ym_r0
6️⃣ Clustering of environmental samples inside the Huanan market where animals were sold.
So far we have shown:
✅ Clear association of cases to the Huanan market
✅ Multiple spillovers likely happened at the Huanan market
✅ Susceptible animals were sold at the Huanan market
But what about inside the market? It's previously been reported that early cases primarily came from the western side of the market (where animals were sold), but anything more specific than that?
Well, yes. Enter China CDC report from January, 2020
Make sure you read it - it's short and has a lot of key details, including location information of the 585 environmental samples they took from the Huanan market.
Their conclusion?
"... it's highly suspected that the current epidemic is related to the trade of wild animals".
The report contains details about the sampling from inside the market. We used this information to reconstruct a detailed map of the Huanan market, overlaying information about live animal sales, environmental sample positivity, and human cases.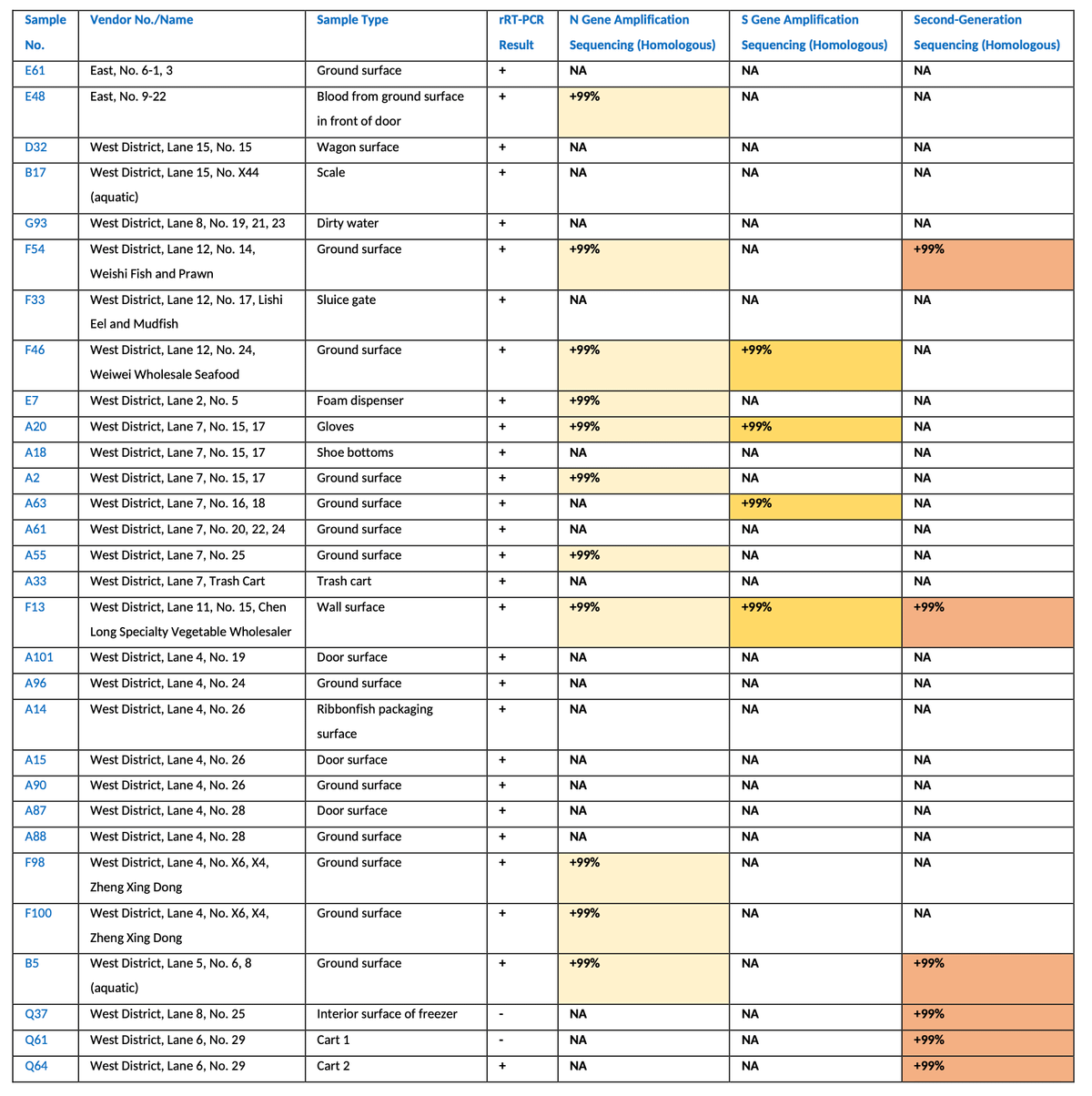
Again, make sure you read the report - and the many other China/Wuhan CDC reports, official news articles, and other relevant material we had translated for our manuscript.
Available here ("File S1"):
zenodo.org/record/6291868
What did we find?
✅ Very clear clustering inside the market
✅ Clustering specifically where live animals were sold
So not only do we find clustering *outside* the market - we find very tight clustering *inside* the market.
Where live animals were sold.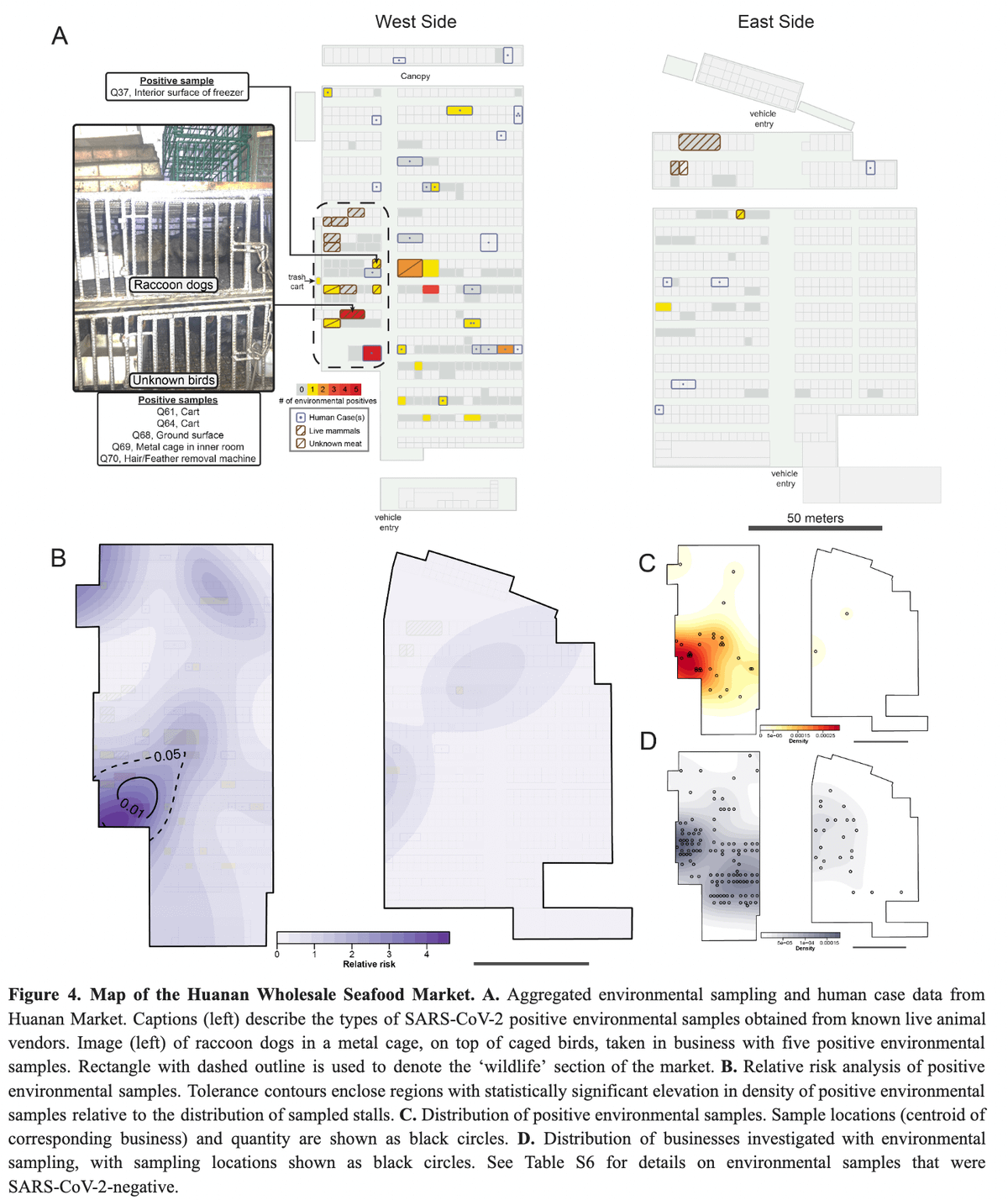
⚠️ Some caveats to these analyses.
Because we didn't perform the sampling inside the market ourselves, we don't have access to all information - most importantly, sampling scheme.
Some specifics on potential bias from @acritschristoph:
twitter.com/acritschristoph/status/1497946228272910337?s=20&t=tXd0UmNZxQN_-mO4qWNOEg
⚠️ The Gao et al. preprint reported that a host read analysis of key raw sequencing data - which we encourage our colleagues in China to make publicly available - showed no animal association.
Some comments from @arambaut on why those analyses are flawed:
twitter.com/arambaut/status/1497696812550918144
7️⃣ Environmental samples clearly associated with animal sales
From the China CDC report we also know what objects positive environmental samples came from.
Most environmental samples (5) came from a single shop that we know sold live animals. What objects were positive?
Well...
✅ Cage
✅ Two carts
✅ Feather remover
✅ Ground
From the Gao et. al preprint we also know:
✅ Sewage outside shop
Here's the crazy thing - co-senior author @edwardcholmes visited the Huanan market in 2014 and took photos of this exact shop! Including raccoon dogs 🤯.
Photos:
Okay, let's have a closer look at these:
✅ Cage
✅ Two carts
Well, cage is clearly there. What about the carts (often used to transport mobile cages and other goods)?
Well, 🔍 .
Two carts in Eddie's photo.
What about:
✅ Feather remover
Well, 🔍 .
The raccoon dogs in Eddie's photo sat on top of birds.
What about:
✅ Sewage outside shop
Well, 🔍 .
The raccoon dogs in mobile cages in Eddie's photo sat on top of the sewage system.
So when we see that the following environmental samples are positive from the China CDC survey:
✅ Cage
✅ Two carts
✅ Feather remover
✅ Ground
✅ Sewage
All those are seen in Eddie's photo - clearly associated with live animal sales.
🤯
And that Lineage A sequence reported in the Gao et al. preprint?
It was located here (figure from @alchemytoday):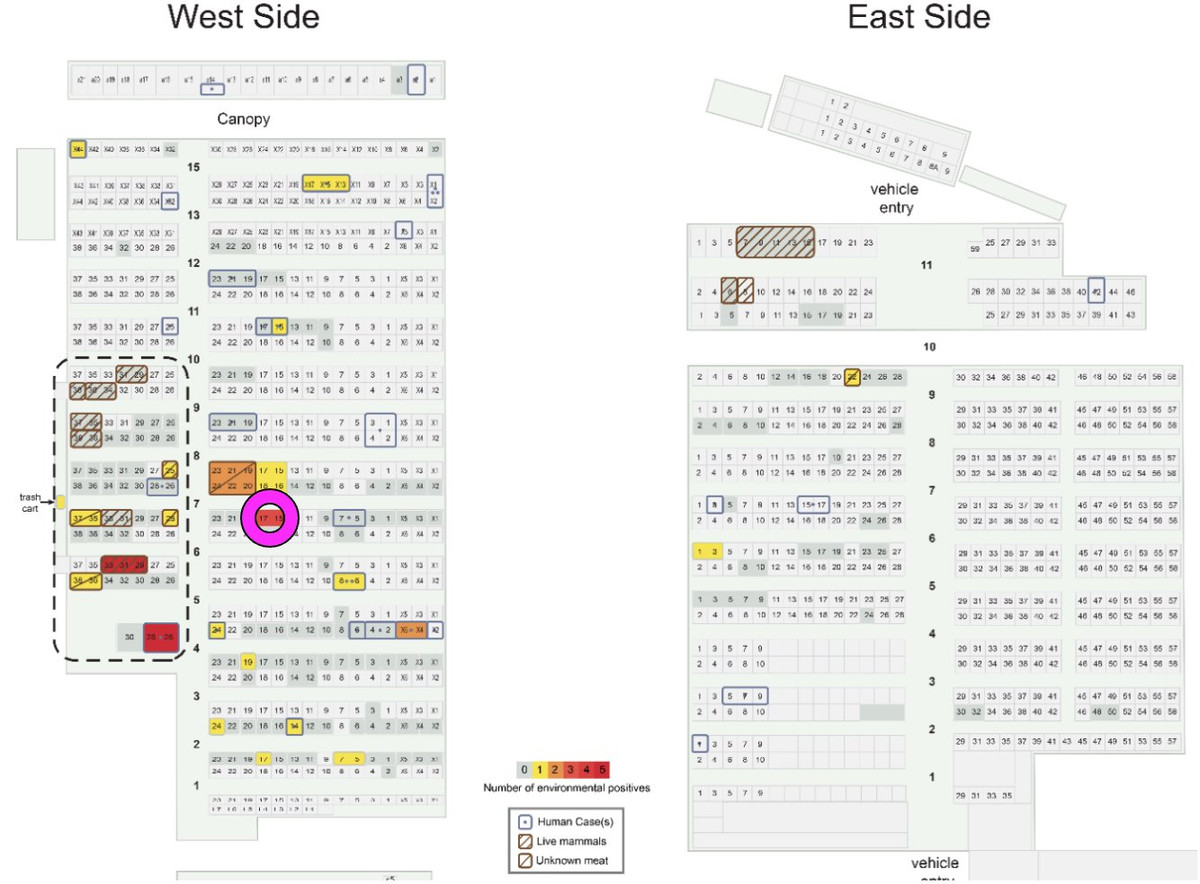
8️⃣ Summary
Combined, all of these data clearly show that the Huanan market was the epicenter of SARS-CoV-2 emergence - with a very likely origin via infected animals.
Other non-market hypotheses are possible, but I don't find any of them plausible. Because, science.
9️⃣ Next steps
So have we solved the origin of the COVID-19 pandemic? No. The data very clearly point to a market beginning via infected animals, however, we have limited understanding of upstream events.
❓What animals?
❓Farmed or wild?
❓Connectedness?
❓Epi?
❓Future risk?
Answering all of these questions will require significant further work and is critical. Tracing back supply chains; genomic epi; serology studies at farms and more widely; in-depth epi/serology studies in Wuhan with a focus on Nov / Dec; ruling out alternative hypotheses, etc.
I'm hopeful that @WHO SAGO - w. China CDC - will help bring further clarity to these very important questions. We need to more fully understand how pandemics start.
To quote @mvankerkhove: Science, solutions, solidarity.
I'll add, collaboration, collaboration, collaboration.
1️⃣ 0️⃣ Extra material
Several excellent resources to explore these studies and questions more.
For example, coverage from:
@nytimes
nytimes.com/interactive/2022/02/26/science/covid-virus-wuhan-origins.html
@washingtonpost
washingtonpost.com/health/2022/03/01/nih-review-lab-safety/
@ScienceMagazine
science.org/content/article/do-three-new-studies-add-proof-covid-19-s-origin-wuhan-animal-market
@Nature
nature.com/articles/d41586-022-00584-8
⭐ threads from co-authors and Tweeps:
@MichaelWorobey:
twitter.com/MichaelWorobey/status/1497607313397481472
@angie_rasmussen:
twitter.com/angie_rasmussen/status/1497692666997329923?s=20&t=l6TLZfiIt2eLazH_b54owA
@whstancil:
twitter.com/whstancil/status/1497971502720901120
@emmecola:
twitter.com/emmecola/status/1498633737411567619?s=20&t=piLqpOQNo2tw30QZje6ylg
@stuartjdneil:
twitter.com/stuartjdneil/status/1498058588253835266?s=20&t=JEFCg4OApygs6HD5RuNk5Q
@PhilippMarkolin:
twitter.com/PhilippMarkolin/status/1498283470627479557?s=20&t=jx0Lw4ifiSLGZ35mIb6vHA
Background information on various aspects of the "origin" question:
@mstandaert on animal farming/sales
washingtonpost.com/world/asia_pacific/china-covid-bats-caves-hubei/2021/10/10/082eb8b6-1c32-11ec-bea8-308ea134594f_story.html
Article on Danielle Anderson who worked at the WIV just as the pandemic started
bloomberg.com/news/features/2021-06-27/did-covid-come-from-a-lab-scientist-at-wuhan-institute-speaks-out
My Q&A with @nytimes
nytimes.com/2021/06/14/science/covid-lab-leak-fauci-kristian-andersen.html
And finally, these two articles:
@janeqiuchina's must-read on Shi Zhengli
technologyreview.com/2022/02/09/1044985/shi-zhengli-covid-lab-leak-wuhan
@stuartjdneil, @past_is_future, and @STWorg on conspiracy theories surrounding the "origin" question:
Finally, if you want to take a look inside the Huanan market @babarlelephant created this 🤯 resource:
babarlelephant.free-hoster.net/visiting-the-wuhan-seafood-market/
You can read the unrolled version of this thread here: typefully.com/K_G_Andersen/718agsO