Like it or not (I don't..), but mass-scale clinical testing for COVID-19 is likely going away in the near future.
Just as we're entering a new wave 📈.
A 🧵 on priority areas I think we *must* focus on:
1️⃣ Wastewater surveillance
2️⃣ Clinical sequencing
3️⃣ Home testing
Important background article from @nature on some of these issues:
nature.com/articles/d41586-022-00788-y
Until now, we have been relying on case and hospitalization data to get a sense of the waning and waxing of the COVID-19 pandemic. This relies on (a) testing being done, and (b) reported.
However, this is likely going away. E.g., 🇩🇰 👇 (NOT 🇩🇰 specific!)
twitter.com/K_G_Andersen/status/1504198260281339905?s=20&t=XIq6AL00xr0UUmBjhMkZOQ
Since testing will get more biased and reporting may likely (mostly) disappear, we're left in the blind on the rise of future waves - until they hit hospitals 🙈 🙉.
Now, we should solve this by *not* decreasing testing, but that's unlikely to happen.
Enter,
1️⃣ Wastewater surveillance
To make up for the lack in testing, we need to heavily invest in wastewater surveillance - measure the amount of SARS-CoV-2 in communal wastewater - across the country. The WHOLE country.
Test and report daily. "Daily" means "every day" btw...
Via our SEARCH Alliance here in San Diego, we're already doing this. But these programs need sustainable funding - which isn't currently the case. We practically have to beg for money on a daily basis - and we're not always successful, leading to delays.
searchcovid.info/dashboards/wastewater-surveillance/
Importantly, this surveillance *must* be combined with sequencing of the wastewater - combined with tools such as "Freyja" that @josh__levy developed in our lab.
This allows us to monitor the dynamics of SARS-CoV-2 variants in wastewater over time. E.g., 👇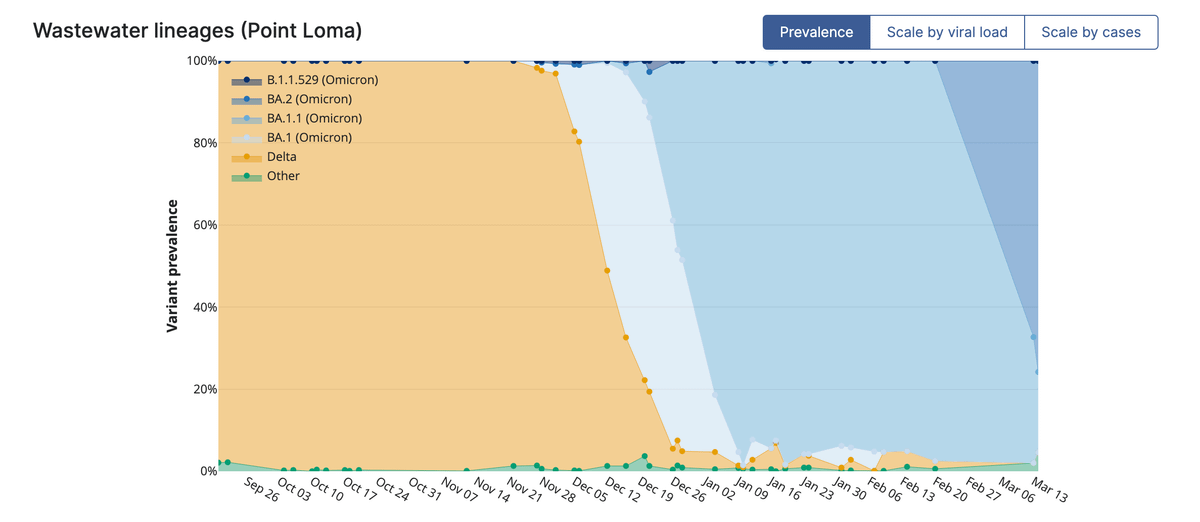
Note, the frequency and speed at which we see lineage displacement = 🤯. We have never observed this with other viruses - so it's key we keep a close eye on SARS-CoV-2 dynamics. This is *not* a process that will magically stop.
Paper on how we do 👆
medrxiv.org/content/10.1101/2021.12.21.21268143v1
Also, here's a link to Freyja (available on Docker, conda, Github, etc).
andersen-lab.com/secrets/code/#freyja
2️⃣ Clinical sequencing
Widescale testing will decrease significantly, which will greatly limit our ability to do non-wastewater based "genomic surveillance" of SARS-CoV-2.
E.g., what SEARCH is currently doing:
searchcovid.info/dashboards/sequencing-statistics/
However, testing of hospitalized COVID-19 patients will presumable continue and it's important to sequence those - and do so consistently.
Why? So we can (a) compare to our wastewater-based sequencing, and (b) if deviations are seen, understand reasons why.
Why might we see deviations? E.g., age-skews in cases, potential differences in variant virulence, etc.
As above, these programs need sustainable support. Do we really need to beg for money for these programs?
SEARCH has been doing genomic surveillance since February, 2020.
3️⃣ Home testing
Finally, we really need to get fully behind testing at home. It simply doesn't make any sense to either (a) go queue forever to get a test - with a result reported days later, or (b) not get a test because they're no longer offered.
This model NEVER made sense.
Because "One size does not fit all" and rapid (antigen) testing is one of the most undervalued tools in our COVID-19 toolbox, @michaelmina_lab and I wrote about it in 2020:
science.org/doi/10.1126/science.abe9187
These tests need to be:
1️⃣ Widely available
2️⃣ Cheap, for those who can afford them and free for those who can't
3️⃣ Ideally, (anonymous) reporting of test results should be included via an app
4️⃣ Positive results should be combined with accessible/affordable treatment options
SARS-CoV-2 will not magically go away because we wish it so - it's "endemic delusion".
Things will get better, but only if we make it so. Wishing away a virus will achieve the opposite - we have more than two years of data and an estimated ~20m dead to underscore that point.
Finally, I want to thank CDC, NIH, and Conrad Prebys foundation for having supported our work so far.
I'm glad to see the CDC is planning on establishing "Pathogen Genomics Centers of Excellence", which I hope can lead to more sustainable 🇺🇸 programs.
cdc.gov/amd/whats-new/pgcoe-nofo.html
You can read the unrolled version of this thread here: typefully.com/K_G_Andersen/8r3HKeN